Tumour Heterogeneity and Cancer Therapy Challenges
Next-generation sequencing is providing new insights into the study of heterogeneous tumours which could help overcome major challenges in cancer therapy. The rapidly-evolving technology allows the examination of the diversity of cells within a tumour. Although initial responses to cancer therapy might be effective, eventual relapse is still possible, and often these new tumours are resistant to the previous therapy. However, understanding the heterogeneity of tumours may reveal optimal cancer treatments for patients, reducing the chance of ineffective treatment or relapse.
In order to understand the mechanism behind this it is necessary to be familiar with the concept of tumour evolution. An unlucky combination of just a few mutations in tumour suppressor genes (genes that have a tumour suppressive function) and oncogenes (genes that have a tumour promoting effect) can be sufficient to initiate tumour formation. As these tumour cells already have an increased turnover rate and the loss of crucial tumour suppressive functions can lead to genomic instability, further mutations are likely to accumulate over time. Cells that carry mutations favouring cell survival and turnover will have a selective advantage and persist in the tumour.
By the time of diagnosis, which can be several years after tumour formation, many mutations may be present in the tumour and subpopulations with different sets of mutations may have evolved. Some subpopulations may be inherently resistant to the selected therapy but the resistant cells might be very rare in the tumour before treatment. Upon treatment, the majority of tumour cells may respond well, leading to a fast regression of the tumour. However, a few resistant cells can be enough to cause a relapse.
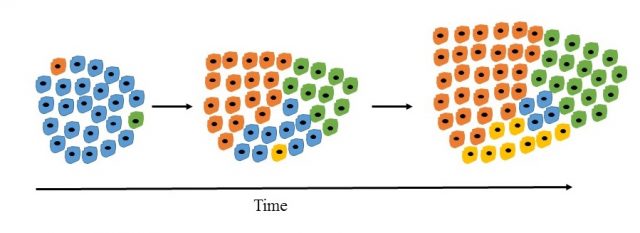
The concept of tumour evolution. Tumour cells acquire additional mutations over time that lead to the emergence of subpopulations. By the time of diagnosis the tumour can be very heterogeneous. Image Credit: Sarah Neidler
It has been thought that these resistance mutations arise during treatment, but there is growing evidence that certain resistance mutations already exist before treatment and give cells a selective advantage upon treatment. For instance, lung cancers with a certain EGFR mutation are sensitive to the EGFR inhibitors Erlotinib. Secondary mutations in the EGFR inhibitor however interfere with the binding of the drug, thereby causing drug resistance and relapse. Close examinations of tumours have shown that these secondary mutations already occur before1. Therefore, a pre-emptive treatment targeting this secondary mutation might be more efficient than treating these cells only after they have expanded due to a selective advantage.
These subpopulations may be very small and therefore difficult to detect, so is there a way to examine all the different subpopulations in an advanced tumour? Next-generation sequencing is a powerful parallel sequencing approach, which allows the detection of genetic alteration events such as mutations, fusion genes and copy number variations. In a recent publication from Silicon Biosystems, small tumour pieces were dissociated and cells were brought into suspension2. Specific antibodies were then used to visualize certain surface markers present on different populations of cells, in this case tumour cells and non-tumour cells.
The research team then used DEPArrayTM technology to obtain pure cell populations. The DEPArrayTM system allows individual cells to be manually selected on a computer screen for further analysis. Because the cell population is not contaminated by non-tumour cells such as infiltrating immune cells, only a few hundred cells are sufficient to obtain clear results about the mutations that are present in this population. With this approach, many different tumour regions can be analysed and antibodies can be used to distinguish different cell populations.
Mutations that are found throughout the tumour are likely to be an initiating driver mutation and these mutations are promising targets for therapy. Moreover, mutations that give the cells intrinsic resistance to a certain therapy can be identified this way and will facilitate finding the optimal treatment option.
Edited by Sarah Spence
References
- This link leads to the study that shows that the secondary T790M mutation occurs already before treatment: http://cancerres.aacrjournals.org/content/66/16/7854.long
- In this publication DEPArray technology was used to obtain pure cell populations for Next-Generation-Sequencing: http://www.nature.com/articles/srep20944










