Tackling the mysterious 8%: A new chapter for human DNA

In almost every single one of the ~37 trillion cells in the human body1, there exists a simple (genetic) code made up of four chemical units. This code, called deoxyribonucleic acid or ‘DNA’, is a combination of these four units (‘bases’):
Adenine (A), Thymine (T), Cytosine (C), and Guanine (G).
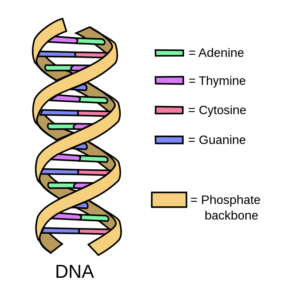
Adenine (A), Thymine (T), Cytosine (C), and Guanine (G). The DNA double helix structure. Image by Forluvoft (CC BY 2.0)
Most of the DNA in a cell is stored as structures called chromosomes which can be found inside the nucleus. In fact, about 2 m of DNA is twisted up into a tiny space of around 10 µm in diameter! 2. DNA itself is made of these four bases (A, T, C and G) chained together like a twisted up ladder which scientists call a DNA double helix. The two sides of the ladder are joined by chemical interactions between these bases: A pairs with T and G pairs with C3. To be precise, we know about ~ 3 billion of these bases are paired up inside each human cell; collectively called the ‘genome’4.
Hidden within our DNA is an astonishing amount of instructions used to make a human being from scratch. And, at the simplest level, depends upon these 4 DNA bases. From eye colour5 to the shape of your ears, it’s all in there somewhere. However, some of the information hidden away in our genetic code may also tell us a lot about diseases of the human body. For example, why some individuals become sick with diseases like cancer and others don’t6.
But to really understand what each instruction does and how instructions go wrong, and in some cases cause disease, we need a way to read and interpret all the information within. Yet it’s only really in the last 20 years scientists have been able to do this, and still about 8% of our DNA code was missing7 until recently. Here, we will investigate the dawn of the human genome and how we have finally (?) completed the human instruction book.
The dawn of the human genome
If we imagine life before the internet, instructions could be found in books stored within libraries; catalogued for you to find what you need. Each of our cells also, like a library, contains a vast amount of information. Up until the 90s, scientists only had some of the instructions, relying on summaries of one line here and there, published for other scientists to read and investigate what the instruction is for. Over time the accessible information expanded, and the known content of the library grew, revealing a vast amount of information. But still, the true view of the library was obscured. Nobody had complete access. And nobody was able to index the contents alone because there was simply so much information. At the time, suitable technologies to do this cheaply and efficiently were still under development. Once the technologies and resources became available, a project was conceived, the core of which centered on a simple question: why not complete the human genome?
And, in 1990, it began. The project which singularly transformed our very understanding of human DNA.
Two groups embarked on tackling this momentous task independently: the Human Genome Project (HGP), a worldwide group of scientists hailing from ~ 20 countries across the globe, and Celera Genomics, a private company originally headed by influential biotechnologist Dr. J. Craig Venter8. Both took different approaches to solve the same problem, and ultimately in 2001, two drafts of the human genome emerged from each consortium, within one day of each other. The HGP published their work in the scientific journal Nature9 and Celera Genomics in the journal Science 10. Approximately 13 years since its conception, the HGP declared completion in 2004, ending over a decade of work, and Celera Genomics a few years earlier. Since their publications, scientists all around the world have extracted the previously untouchable information contained within the human genome library leading to incredible advances in our understanding of what it takes to make a human (at the genetic level). Importantly, and as was hoped, our understanding of how diseases of our DNA, like cancer, develop has rapidly improved. Now scientists can even check for changes to single bases in the DNA sequence and how they may contribute to increased cancer risk 11.
But if our genome was so ‘complete’, then why do it all again?
In reality, our genome was never ‘completed’ in the early 2000s, but was only ‘essentially complete’. To understand why we have to look at how the original genomes were built. During the HGP, scientists cut up human DNA into lots of smaller pieces. These pieces were inserted into bacteria, which grew lots of copies of these different fragments. The fragments were removed from the bacteria and cut up again into even smaller pieces, loaded onto a machine (a sequencer). Then the order of the As, Ts, Gs, and Cs were read for each fragment. Using computers, scientists then stuck all the pieces back together, and bingo, human DNA. Celera genomics used an approach called ‘shotgun’ sequencing. Like the HGP, the DNA was broken up into many smaller pieces. However, these smaller pieces were then not given to bacteria. Instead, they were sequenced on a sequencer, and computers were used to look for overlapping bits i.e presumably they came from the same DNA region. Where they matched, the overlapping bits acted like ‘glue’, sticking the two pieces of DNA together until essentially the whole DNA sequence was finished. Over time, the human genome was updated and updated, improving the quality12. But still, around 8% of mysterious DNA remained; that’s about 300 million unknown DNA bases!13.
These missing regions were problematic. They all looked pretty similar: the same bases in a chain or similar patterns of bases repeated over and over and over again. And though in the 2000s, both scientists and software alike worked tirelessly to resolve ~ 92% of the genome, these repeated regions confounded the best. The problem was the short pieces of DNA that were needed for the sequencing. This meant there wasn’t always enough information around the repeated DNA sequence for either the program or the human to decide exactly where it should go. Unfortunately, some of the missing information was important. For instance, the smaller ‘arms’ of five chromosomes were missing. Scientists were also struggling to understand how the centromeres looked. Centromeres are important because they are the sites on chromosomes that hold together the two pieces (chromatids) that form when the original DNA is copied. They are anchors for the machinery needed to separate the two chromatids when the mother cell divides into two daughters. Some diseases are linked with this process, like Down’s Syndrome (or Trisomy 21), where people have one extra copy of chromosome 2114. Understanding the sequence of this DNA may give clues as to why this happens.
So birthed the Telomere-2-telomere (T2T) consortium, a community of scientists, co-led by Drs Karen Miga and Adam Phillippy, set on uncovering the true nature of these missing regions and (finally) completing the genome from one end of each chromosome (the telomere) to the other15. Their work, unlike the HGP and Celera Genomics, was made possible by rapid advances in sequencing technologies and better software.
Nowadays, some sequencing machines can sequence longer and longer pieces of DNA. These ‘long-reads’ i.e very long pieces of uninterrupted DNA sequence, are much easier to work out which bit overlaps with what as there is simply more information around mysterious repeating regions to make the decision; effectively solving the ‘repeat’ problem. Using this approach, the T2T consortium, on the final day of March 2022, published the complete sequence of a human genome in Science. A final total of 3.055 billion base pairs with almost no gaps16.
What did they find?
This new genome (“T2T-CHM13”) now contains the missing information: 5 chromosome arms, centromeres, and telomeres (sequences of DNA to protect the ends of chromosomes). All of which are very repetitive regions of our DNA. Even in the short space of time between the genome being published and now, thousands of potential avenues to investigate further have opened. Some of these previously missing pieces may aid in how the body defends itself against infections, why we have big brains, developed brains and there may even be new ways we could target cancer hidden amongst the many repeated DNA bases18 . Interestingly, this new genome may even help us understand better how humans age. Current research suggests longer telomeres are associated with longer life. And now, with improved telomere sequences, scientists can delve deeper into the roles of these important DNA structures19.
What surprised many, however, was the lower number of core instructions (or ‘genes’) than previously thought were needed to make a human. This may mean that though we use a comparably smaller core instruction set that forms the foundation, more complex alterations, and interactions, occurring in ways we do not yet understand, may really shape who we are and what our bodies become. Continuing work on the human genome will reveal more in the coming years.
So now the genome is complete. Are we done yet?
No.
This DNA sample came from a cell with one set of chromosomes (‘haploid’), forming when something went wrong. When egg and sperm met, a problem occurred, keeping only the sperm cell’s information. In this case, the sperm cell was carrying 23 chromosomes (humans have 46 or 23 pairs) including an X-chromosome. Genetically male humans have one X chromosome, and another called Y. The published genome misses both a complete T2T Y-chromosome and what is happening on the paired chromosomes. In reality, this genome is just a single picture. It comes from a special type of cell, at one point in time and space, from an individual that never came into existence. Though it is representative of humans, we know that DNA can change a lot over time and will vary from one person to the next.
The task for scientists now is to investigate what parts of this sequence represent us all and which parts don’t, but we need complete genomes, sequenced T2T, from a diverse population of humans across all areas of our vast planet to really understand this.
And what about genetic diseases, like cancers? What is the same and what is different? How can we apply this knowledge to make better decisions in the medical field for more effective, safer, and maybe even more personalized treatments? Will it be possible one day to make a drug just for you? Could we use our genomes routinely to find diseases before they start? Can we delay aging?
With technology becoming cheaper and faster, it could be possible in the not-too-distant future for everyone to have their genome sequenced and their library indexed. But what if we do find what affects intelligence, personality, cancer predisposition, and genetic disease? How is this information stored? Who do we share it with? Can we deal with knowing ourselves and others on the genetic level? What happens when we find there is no treatment?
That said, what is so exciting about this new genome is exactly that-The unknown. We are left with so many positive opportunities to advance our understanding of ourselves.
Beginning with the stories missed over the 20 years.
Specialist-edited by Shona Richardson
Copy-edited by Sridevi Kuriyattil
References
- https://commonfund.nih.gov/HuBMAP
- https://www.sciencefocus.com/the-human-body/how-long-is-your-dna/
- https://www.genome.gov/genetics-glossary/Deoxyribonucleic-Acid
- https://www.nature.com/scitable/topicpage/dna-sequencing-technologies-key-to-the-human-828/
- https://www.verywellhealth.com/genetics-of-eye-color-3421603
- https://www.cancer.gov/about-cancer/causes-prevention/genetics
- https://archive.nytimes.com/www.nytimes.com/library/national/science/062600sci-human-genome.html?amp;sq=francis%252520collins&st=cse&scp=23
- https://www.nature.com/scitable/topicpage/dna-sequencing-technologies-key-to-the-human-828/
- https://www.nature.com/articles/35057062
- https://www.science.org/doi/10.1126/science.1058040
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5746410/
- https://www.nature.com/scitable/topicpage/dna-sequencing-technologies-key-to-the-human-828/
- https://www.science.org/doi/10.1126/science.abj6987
- https://www.nature.com/scitable/topicpage/trisomy-21-causes-down-syndrome-318/
- https://sites.google.com/ucsc.edu/t2tworkinggroup
- https://www.science.org/doi/10.1126/science.abj6987
- https://edition.cnn.com/2022/03/31/health/first-complete-human-genome-sequence/index.html[/note] 17https://www.sciencefocus.com/news/first-complete-human-genome-reveals-genetic-variants-disease/
- https://www.frontiersin.org/articles/10.3389/fgene.2020.630186/full