Since the birth of modern medicine, research has provided extensive knowledge about the biological events leading to complex diseases such as cancer. Benchside science continues to enable the development of effective drugs which target the root cause of the disease, known as targeted therapy. Whilst targeted therapy has the advantage of minimising unwanted side effects, doctors must be able to predict which patients will respond to a given class of drug. Fortunately, recent genetic studies have shown a patient’s response to treatment may be written in their DNA. Particularly within the field of cancer research, scientists and clinicians have been forced to embrace a fundamental shift from a traditional “one size fits all” approach to that of precision medicine, which describes a model in which an individual’s treatment is directed by subtle clues, or mutations, in the individual’s unique genetic code, or DNA.
The development of personalised medicine began with a stratified approach, which aimed to treat patients according to their subtype of disease, as opposed to their individual genetics. Clinically-distinct subtypes of breast cancer have been defined since 2012 (TCGA) and have been shown to drastically improve treatment outcomes of patients with this disease. One subtype known as HER2-positive breast cancer describes the presence of the human epidermal growth factor receptor-2 (HER2) protein on tumour cell surfaces. Although this subtype only accounts for 1 in 5 breast tumours, this finding allowed doctors to prescribe Herceptin, a targeted therapy which specifically blocks HER2, inhibiting a known cellular pathway which otherwise enables cell growth and proliferation. Therefore, Herceptin successfully reduces tumour growth and induces cell death in a defined subset of patients whilst minimising the severe side effects of traditional chemotherapy.
Recently, advances in genetic profiling have provided scientists with a means to interrogate a high volume of DNA samples from patients with all kinds of cancer. Although, like in breast cancer, this has led to increased understanding of disease subtypes in pancreatic tumours, clinical management of the latter has remained unchanged for 40 years. Unfortunately, scientists have yet to successfully target the main genetic faults in pancreatic cancer due to the highly variable, or heterogeneous, genetics of this disease. Therefore, scientists aim to identify clues that guide a combination approach to treatment, using a lower dose of traditional chemotherapy alongside a less toxic, targeted drug. Most recently, scientists have started trialling a drug known as Olaparib in the 2% of pancreatic cancer patients that possess a fault in the DNA sequence, or gene, known as BRCA1. Although a fault or mutation in BRCA1 is not a main ‘driver’ of pancreatic cancer, scientists have already shown that these patients are likely to benefit from olaparib which targets a related, ‘sister’ protein, PARP1. Olaparib, an inhibitor of PARP1, is an example of a targeted drug which has been shown to improve overall survival in breast cancer and ovarian cancer patients, prior to testing its utility for the treatment of pancreatic tumours.
Due to the relatively low frequency of targets, such as the BRCA mutation or the presence of HER2 in pancreatic cancer, it is difficult for scientists to justify creating a dedicated clinical trial for such drugs. This has meant that scientists, both in Glasgow and beyond, are required to redesign clinical trials that we hope will identify genetic flags which will predict which treatment is the best for patients. Here in Glasgow, clinicians and scientists have developed a series of clinical trials under the title, Precision-Panc. This umbrella trial will look at the utility of known drugs in the context of pancreatic cancers (such as those that target faults in DNA repair pathways, similar to BRCA). In addition, these trials hope to improve our understanding of which patients respond best to either of the current chemotherapeutic regimes, FOLFIRNOX or Abraxane/Gemcitabine. One way to investigate differences in drug response between patients is to replicate a patient’s tumour in the lab and test for the most effective drug combination prior to performing genetic tests to identify genetic flags known as biomarkers which correspond with a better drug response.
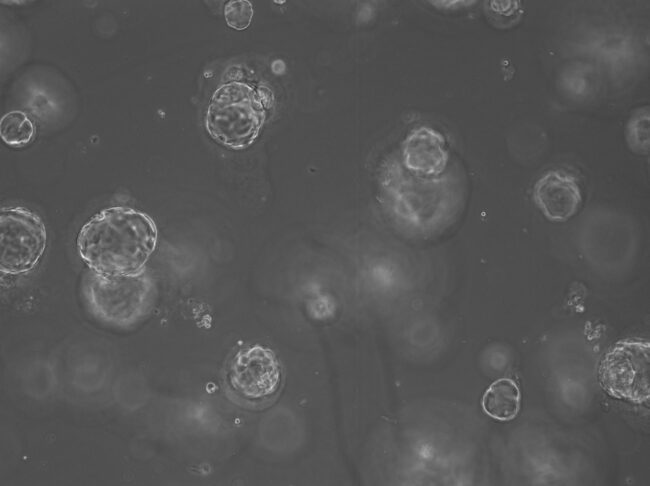
This approach to personalised medicine has already been addressed by world-leading scientists, who continue to develop 3D cellular models known as organoids, or “mini-organs” from small amounts of patient tissue. Such models of disease are particularly advantageous within the field of cancer research due to their ability to recapitulate a patient’s own genetics and accurately predict response to a proposed drug regime. Unlike 2D models, organoids mimic physiological structures and allow the study of drugs on different cell populations within a tissue. Organoids maintain their genetic characteristics over time in a Petri dish (unlike 2D cultures) and can be frozen for future use in later studies. Most excitingly, scientists continue to report success growth of organoids from all tissue types including colorectal, prostate, pancreatic and breast tumours. In recent years there has been a national effort to produce organoid biobanks as a tool for researchers and clinicians. Organoids may assist the translation of precision medicine into clinical practice in two major ways. However, the best way to utilise these “mini-organs” is not defined.
On one hand, studies have shown that organoids can be used in functional tests which involve treating a patient’s organoid with a drug prior to clinical administration of the best drug. However, scientists predict that using organoids for drug screening could require up to 3 months to simply grow enough ‘mini-organs’ for high volume testing, which is not suitable for diseases that progress quickly, such as pancreatic cancer. Otherwise, organoid technology can be used to allow deeper sequencing of tumour cells, enabling low-frequency mutations, such as BRCA, to be identified. Deep sequencing is not often directly available from tissue samples, as these are often very small and therefore provide suboptimal amounts of starting material for complex genetic tests. Furthermore, experts believe that we may be looking for a pattern of faults across multiple genes, or gene signature, to identify those patients that will respond well. Therefore, scientists need to be able to test for hundreds of genes simultaneously which requires sufficient volumes of DNA. By combining genetics, derived from a patient’s organoid and cross-referencing this with clinical outcomes or treatment response, scientists hope to help identify patterns in genetic faults which will then guide future clinical decisions regarding a patient’s treatment options.
Despite organoids still being relatively novel, scientists already hope to update the current ‘mini-organ’ model, by introducing immune cell components into these 3D structures, allowing the creation of an advanced co-culture model. By introducing a patient’s own immune cells to their respective organoid, scientists could test whether immunotherapies, or drugs that aim to improve the immune system response to cancer, may be advantageous for subsets of patients. Advanced 3D models would also enable studies into unknown mechanisms of drug resistance, which remains a major challenge in drug development across all medical research. Overall, personalised medicine aims to provide the right drug, to the right patient, at the time and scientists hope to implement such an approach in the clinical management of devastating illnesses, such as cancer, neurological disease and immunological defects for which few treatment options are available.
This article was specialist edited by Salvia Novianti and copy-edited by Raeesah Maqsood.